___________________
CRISPR/CAS9, a new laboratory tool
The CRISPR/CAS9 technology (Nobel Prize in Chemistry 2020) is revolutionary in the “molecular scissors” category, thanks to its simplicity and low cost of implementation. Guided by its associated RNA, the CAS9 endonuclease specifically cuts the targeted DNA sequence (region of interest). Double-strand breaks are usually resolved by non-homologous end joining (NHEJ), generating indels (knockouts of the targeted gene). By transfecting a template DNA in parallel with the system expressing CAS/guide RNA, the cut DNA can be repaired in a controlled manner by homologous recombination, or even modified (point mutation, myc tag insertion, reporter gene fusion, etc.).
In the laboratory, we have set up the biological tools needed to use this fast-growing technology:
* We use an expression plasmid for the eSpCAS9 protein (high-fidelity CAS9), into which any target sequence can be cloned so that transfected cells synthesize eSpCAS9 and a specific guide RNA: in this way, any gene of interest can be targeted for “cutting”.
* For “controlled” repair with GFP insertion, for example, we construct a template plasmid containing the sequence encoding GFP, flanked by regions homologous to the targeted region (around the CAS9 cut site). An antibiotic resistance gene (Puromycin or Blasticidin) is included in the insert to select edited clones.
Here are two examples of successful insertion of the GFP sequence into the genome of cultured mouse cells, using the CRISPR/CAS9 technique:
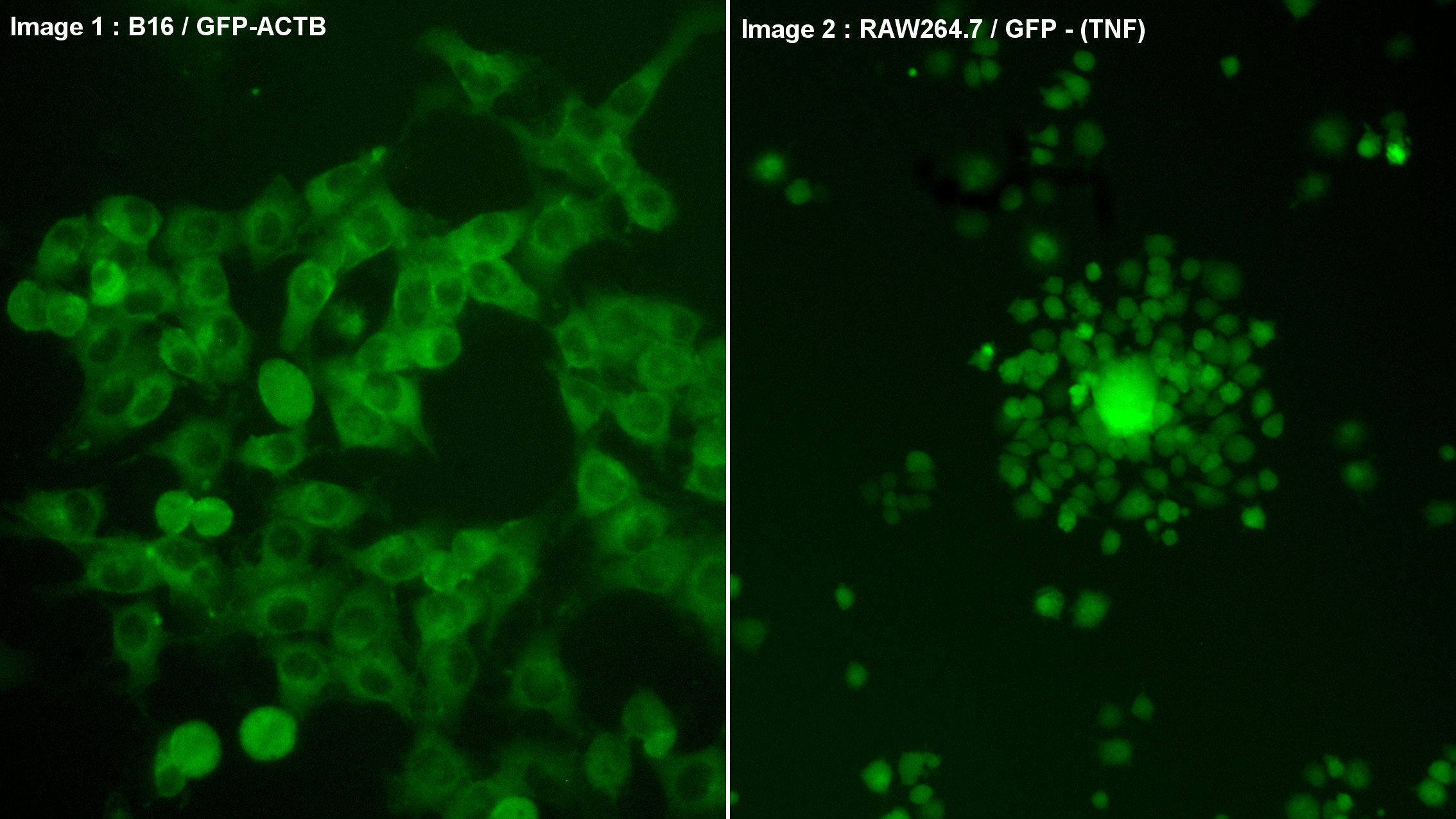
Image 1: Skin cells constitutively expressing GFP-Actin-β, which fluoresces the cellular network of filaments.
Image 2: Macrophages producing GFP fluorescent protein after activation of the TNF-α promoter by LPS
This technology is now available in several “non-cutting” variants in which CAS9 is deactivated (dCAS9) and coupled to various effectors, opening up new possibilities for action on genomic DNA (epigenetic modifications, gene activation/inhibition).
_________________
Also to be read

October 2025 – Thesis defense at UTCBS: Mitta PIERRE
On October 1st, 2025, Mitta PIERRE successfully defended her doctoral thesis entitled “Nanoformulations of antioxidant active ingredients for ophthalmic administration in the prevention of age-related macular degeneration (AMD)” supervised by Pr. Christine CHARRUEAU and co-supervised by Dr. Diana LAMAA
September 2025 – [Keynote research] Nathalie Mignet,Head of Laboratory, Nanomedicines and lipid nanoparticles for nucleic acid delivery
[Keynote research] Nathalie Mignet, Nanomedicines and lipid nanoparticles for nucleic acid delivery

July 2025 – Workshop du master erasmus mundus Nanomed
Workshop du master erasmus mundus Nanomed

June 2025 – Université Paris Cité’s Apprentis Chercheurs conference was held last June, with UTCBS taking part.
Université Paris Cité’s Apprentis Chercheurs conference was held last June, with UTCBS taking part.
